Abstract
Granulocyte colony-stimulating factor (G-CSF) is generally recommended to hasten the recovery of neutropenia after autologous hematopoietic stem cell (HSC) transplantation. However, in patients pre-treated with chemotherapeutic agents, G-CSF was shown to exacerbate HSC toxicity triggered by these drugs by promoting their differentiation or senescence. We reasoned that G-CSF might also negatively impact HSCs treated with DNA-damaging programmable CRISPR/Cas9 nucleases and potentially reduce their engraftment in vivo. To address this question, we previously treated human CD34+ cells by electroporation of AAVS1-specific sgRNA/Cas9 ribonucleoprotein complexes ('RNP group') or Cas9 alone ('Cas9 group'), and transplanted cells from each group into NSG recipient mice. To model conventional autologous transplantation procedures, animals received a daily subcutaneous injection of G-CSF or a control saline (PBS) solution for 14 days post-transplantation. The use of G-CSF post-transplant significantly impaired long-term engraftment of CRISPR/Cas9 gene-edited HSCs (Araki, ASH 2021). In this study, we interrogate at the single-cell level the molecular mechanisms underlying the G-CSF-mediated engraftment defect of gene edited HSCs and provide an approach to overcome the functional impairment.
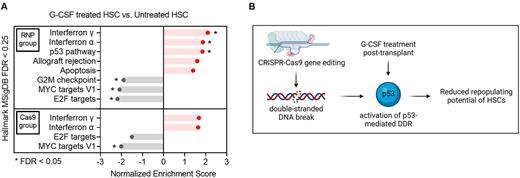
We analyzed human CD34+ cells harvested from the animals' bone marrow after G-CSF or PBS treatment using Cellular Indexing of Transcriptomes and Epitopes sequencing (CITE-seq). We identified 7 distinct clusters (clusters 0 to 6) in dimension-reduction analysis. HSCs were computationally assigned to cluster 6 by associating cluster-specific transcripts with HSC signatures and by comparing CITE-seq CD38, CD45RA, CD90, CD49f expression. Gene set enrichment analysis (GSEA) of cells within the HSC cluster revealed a significant upregulation of interferon α/γ response pathways in G-CSF treated HSCs in both RNP and Cas9 groups, indicating that G-CSF stimulated an inflammatory response in engrafted human HSCs in-vivo independent of the gene editing process (Fig A). In contrast, significant G-CSF-mediated up-regulations of the p53 pathway, a central network orchestrating DNA-damage response (DDR) mechanisms, and of the cellular apoptotic response were observed only in gene edited HSCs (Fig A). In addition, the G2-M DNA damage checkpoint, a key cell cycle regulator that prevents entry into mitosis until damaged DNA is sufficiently repaired, was exclusively downregulated in the RNP group by G-CSF treatment (Fig. A). To further substantiate the impact of G-CSF on gene edited HSCs, we transiently inhibited the p53 pathway by co-electroporating GSE56 mRNA during gene editing ('RNP/GSE56 group'), the approach previously shown to improve graft function of gene edited HSCs (Schiroli, Cell Stem Cell 2019). Addition of GSE56 mRNA offset the transcriptional differences previously observed between G-CSF treated and untreated gene edited HSCs. Next, to evaluate whether the transcriptional changes induced by GSE56 altered engraftment of gene edited LT-HSCs in vivo, we transplanted CD34+ cells electroporated with RNP/GSE56 into NSG mice and injected G-CSF or PBS once daily for the first 14 days after cell infusion. Notably, we observed a comparable long-term (22 weeks) human cell engraftment between G-CSF treated and untreated mice, suggesting that transient p53 inhibition ameliorated the negative effects of G-CSF on gene edited LT-HSC function.
In conclusion, our data indicate that G-CSF administration post-transplant impairs long-term engraftment of CRISPR/Cas9 gene-edited HSCs by exacerbating the p53-mediated DDR triggered by Cas9-mediated double stranded DNA breaks (Fig B). Perturbation of the G2-M DNA damage checkpoint induced by G-CSF may further contribute to the molecular mechanisms underlying the engraftment defect of gene edited HSCs. As clinical trials are implemented, the potential for G-CSF to exacerbate HSC toxicity mediated by DNA-damaging nucleases should be considered.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal